Description

The Snakemake workflow management system is a tool to create reproducible and scalable data analyses.
Software Category: tools
For detailed information, visit the Snakemake website.
Available Versions
The current installation of Snakemake incorporates the most popular packages. To find the available versions and learn how to load them, run:
module spider snakemake
The output of the command shows the available Snakemake module versions.
For detailed information about a particular Snakemake module, including how to load the module, run the module spider command with the module’s full version label. For example:
module spider snakemake/9.8.1
| Module | Version |
Module Load Command |
|---|
| snakemake | 9.8.1 |
module load snakemake/9.8.1
|
Snakemake workflow:
- Snakemake is a workflow management system used to create reproducible and scalable data analyses
- Workflows are written in Python and can be deployed in parallel on the HPC system
- Workflows can be executed in containerized environments: Conda or Apptainer
Snakemake rules:
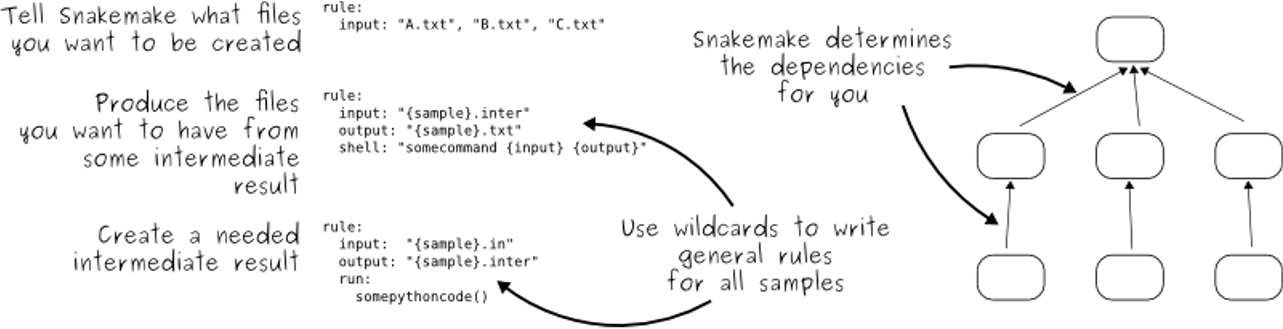
- Snakemake follows the GNU Make paradigm
- Workflows are defined in rules, starting with the target rule
- Dependencies between the rules are determined automatically, creating a DAG (directed acyclic graph) of jobs that can be parallelized
Config.yaml file:
- Config files are for users to input filenames and paths for the workflow
- In the case below, the user inputs 3 samples for a simple RNA-seq pipeline
- Threads can be passed as an argument for multithreading
#Inset sample names as a list:
SAMPLES: ["exp1", "exp3", "Sham1"]
#Insert path to GTF file:
GENOME_GTF: "refGene.gtf"
#Insert path to Hisat2 index files:
HISAT2_INDEX_PREFIX: "grcm38_snp/genome_snp"
#Insert the path to the directory that has your fastq files:
DATA_PATH: "/project/some_directory/fastq_files"
#Each sample should have R1 and R2 extensions:
R1_EXT: "_R1_001_paired.fq.gz"
R2_EXT: "_R2_001_paired.fq.gz"
#Insert number of threads you would like to use:
THREADS: 8
Snakefile:
- The Snakefile contains the rules of your workflow (the steps)
- The target rule is your final output, Snakemake will determine the order of the rules in order to create that output
- Each rule consists of 3 required parts: the input files, the output files, and the shell (command)
- Below is an example of a rule to align sequences using hisat. The log and threads options are optional, but included for reference
- The target output is a gene count matrix in a csv format
#Target Rule (last output):
rule all:
input: "differential_expression/gene_count_matrix.csv"
rule align_hisat:
input:
fq1= config['DATA_PATH'] + "{sample}" + config['R1_EXT'],
fq2= config['DATA_PATH'] + "{sample}" + config['R2_EXT'],
hisat2_index=expand(f"{HISAT2_INDEX_PREFIX}.{{ix}}.ht2", ix=range(1, 9))
output: "align_hisat2/{sample}.bam"
log: "align_hisat2/{sample}.alignment.summary"
threads: config['THREADS']
shell:
"hisat2 2>{log} -p {threads} --dta -x {HISAT2_INDEX_PREFIX} "
"-1 {input.fq1} -2 {input.fq2} | "
"samtools sort -@ {threads} -o {output}"
- After the rule
align_hisat is completed, the workflow can move to the next rule stringtie_assemble
- Notice that the output of
align_hisat is a .bam file, this is now the input to the rule stringtie_assemble
rule stringtie_assemble:
input:
genome_gtf=config['GENOME_GTF'],
bam="align_hisat2/{sample}.bam"
output: "stringtie/assembled/{sample}.gtf"
threads: config['THREADS']
shell:
"stringtie -p {threads} -G {input.genome_gtf} "
"-o {output} -l {wildcards.sample} {input.bam}"
- You can add as many rules as you like as long as they are sequential with inputs and outputs
Slurm for Snakemake:
- The Snakemake pipeline can be executed using a
SLURM script on the HPC system
- Below is an example script to submit to the standard partition with 8 threads
- This script is using a
conda environment called rnaseq
#!/bin/bash
#SBATCH -t 5:00:00
#SBATCH -p standard
#SBATCH --mem=50000
#SBATCH -A allocation_name
#SBATCH -n 9
module purge
module load miniforge
source activate rnaseq
snakemake -p -j 8
Dry Runs:
- Dry-runs are a great way to check your commands before running them
- The code is printed, but not actually run
- For a dry run, use
snakemake -n
|
HPC, software, bio
multi-core